Sequence Dot Plots part II
In the last post, we built a simple WPF application to display a dot plot – this is a graphical representation to compare two streams of data against each other to determine similarity. It is often used in bioinformatics to compare genomic sequences and determine if they are related in some fashion. Comparing the same sequence against itself (obviously has a high degree of similarity) generated a lot of false positives in our application -
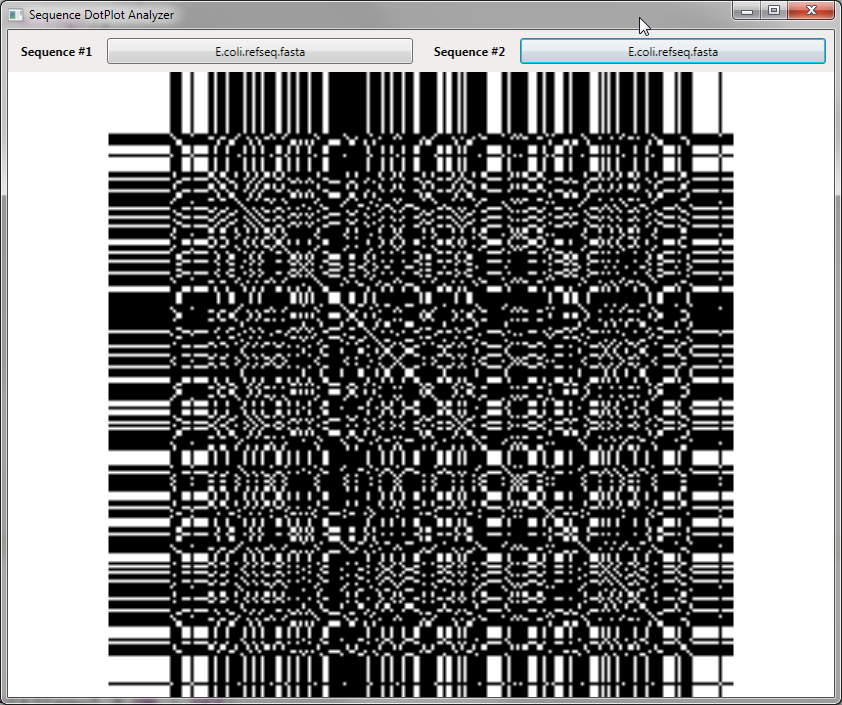
Here you can see the white diagonal line – this represents where the nucleotide data matched perfectly, however we can also see a lot of other white areas where the 4-character alphabet also happened to match at that particular position. Given our small alphabet we have a possible 1 in 4 chance of a match at any particular position (this is perhaps not completely accurate – biological sequences often have specific pairings so it’s clearly not random data but still the character set size certainly has a high probability of matches).
I mentioned that our goal would be to fix the above mismatches. There are a couple of ways to do this – we’ll take the easiest approach which is to increase the required length of the match before we consider it a match – in order words, before we color a pixel white, we must check both before and after that position and ensure adjacent nucleotides also match.
To start with, let’s assume a 3-character match, i.e. both sides of the position must also match. We’ll break out our match test to a method and write it like this:
private bool IsMatch(long s1, long s2)
{
long t1 = s1 + 1, t2 = s2 + 1;
long r1 = s1 - 1, r2 = s2 - 1;
if (r1 < 0 || r2 < 0 || t1 >= _sequences[0].Count || t2 >= _sequences[1].Count)
return _sequences[0][s1] == _sequences[1][s2];
return _sequences[0][r1] == _sequences[1][r2]
&& _sequences[0][s1] == _sequences[1][s2]
&& _sequences[0][t1] == _sequences[1][t2];
}
Given our two sequences (_sequences[0] and _sequences[1]) we will test a specific position in each sequence (s1 and s2) and also adjacent indexes in the sequences. When we run with this implementation we get the following output:
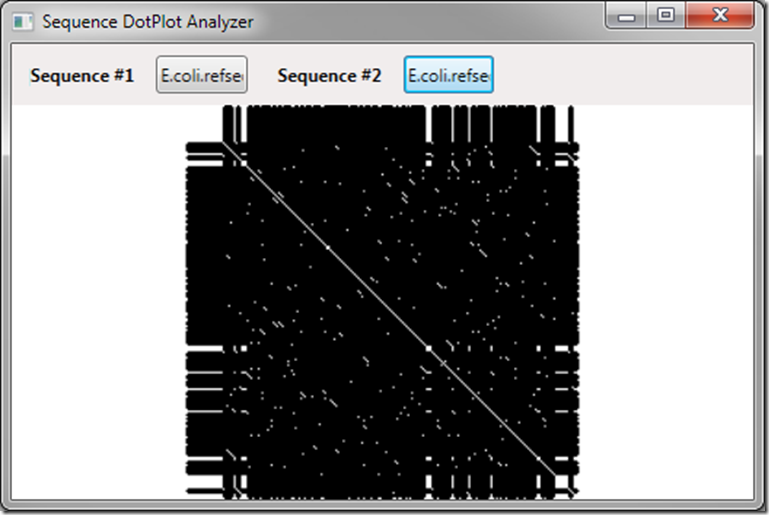
This actually cleans up our data quite a bit – notice the diagonal line is now very distinct but we still have pockets of incorrect matches. We can generalize our method – using a changeable field to determine the window size required for a match – we’ll call the field _requiredLength and expose it with a public property we can data bind to with a slider so we can change it:
private bool IsMatch(long s1, long s2)
{
int match_length = (int) _requiredLength;
if (match_length <= 2)
{
return _sequences[0][s1] == _sequences[1][s2];
}
bool hasMatch = true;
for (int add = -1 * match_length / 2; hasMatch && add <= match_length / 2; add++)
{
long t1 = s1 + add;
long t2 = s2 + add;
if (t1 < 0 || t2 < 0 || t1 >= _sequences[0].Count || t2 >= _sequences[1].Count)
continue;
if (_sequences[0][t1] != _sequences[1][t2])
hasMatch = false;
}
return hasMatch;
}
Then we will add the proper UI bits in XAML and run the application again with a slider to change the value at runtime:
<StackPanel>
<Slider Value="{Binding RequiredLength}" Minimum="1" Maximum="50" Width="100" SmallChange="1" />
<TextBlock Text="{Binding RequiredLength, StringFormat=N0}" HorizontalAlignment="Center" />
</StackPanel>
Sliding the slider will increase/decrease our required length – at zero we get just a single element comparison and it has the original output. When we increase up to 40 – we see a much more evident match set:
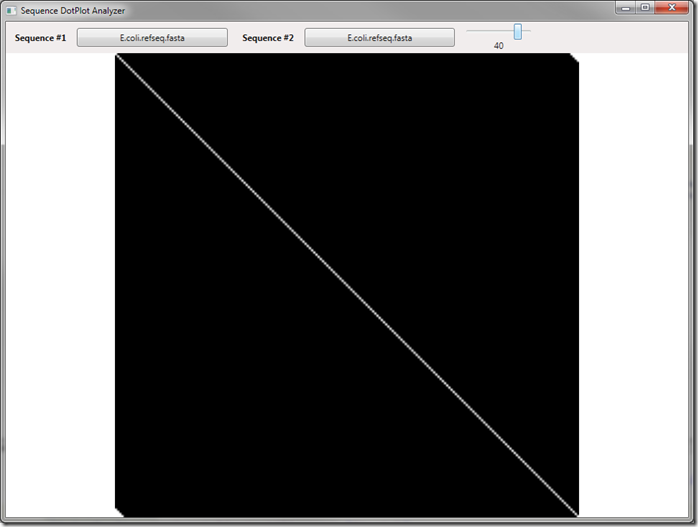
Notice that now our proper match is extremely clear, using this application to check completely unrelated sequences also is useful – we see a complete black screen once we get above a window size of 4 or so. This technique could be used to compare other types of data as well – it’s a versatile and powerful visual way to check data!
The final project is here if you’d like to try it for yourself!