WPF + Prism + Biology!
I spend a lot of time in WPF – both in teaching it around the world, and also in assigning in development of applications for various companies. I also work a bit in the Bioinformatics space – doing applications and research in genomics. This post is the start of a series which will combine these efforts and attempt to give others some helpful ideas on doing bioinformatics with .NET – and creating visualizations with WPF!
.NET and Biology?
To start out, let me introduce a somewhat unknown library from Microsoft Research that I’ll be using – .NET Bio (http://bio.codeplex.com). .NET Bio is actually several things bundled into one. It is a set of usable command line tools which perform tasks such as alignments, assembly and file conversion. It is also a reusable class library that has support to program common bio tasks using .NET. It is this latter area that I want to tackle first, although we might delve into the tools periodically too. It’s a bit primitive in places, but a great foundation to build upon, and highly extensible so if it doesn’t have what you need, it’s fairly simple to add it yourself.
Prism
Prism (http://compositewpf.codeplex.com) is a library for build composite UI applications using WPF. It’s owned and maintained by Microsoft Patterns and Practices (P&P) and essentially gives you the ability to create complex applications by combining various bits and pieces together. These pieces can interact with each other without being tightly coupled – which is great for larger, enterprise style applications, or applications where you plan to extend the features. It allows you to define your UI in terms of “regions” where you plug in various UI elements at runtime. These UI modules are defined independently of the main application structure – and are dynamically composed into the UI and can access and even expose services for other modules to consume. There was a nice overview of Prism in a past issue of MSDN – http://msdn.microsoft.com/en-us/magazine/cc785479.aspx if you are interested in more details on the framework.
Ok, so what’s first?
I was recently playing with Prism, and wanted to see if I could build something extensible that showed off Prism but also showed off the Biology features of .NET Bio – without being too complex to understand. Here’s what I came up with:
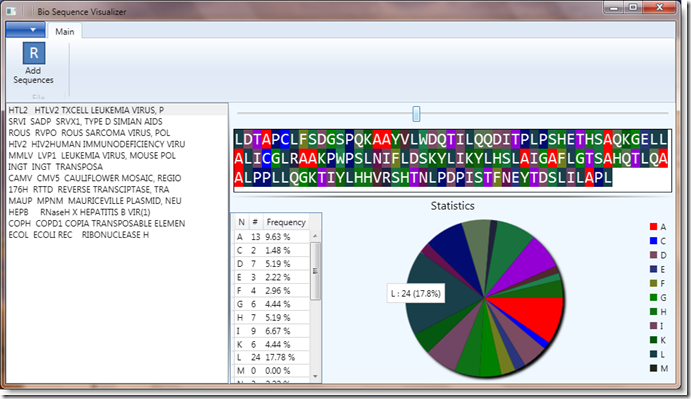
This is the SequenceAnalyzer. I plan to add additional features to it over time, but for now it has a simple feature set:
- It loads all supported file types.
- It displays the loaded sequences from files in a list and allows you to select one
- It shows the sequence data (with coloring applied)
- It shows the statistics (frequency count) from the selected sequence
In addition, it performs the above in a decoupled fashion, using different modules for each piece. It has three basic modules:
- Ribbon
- Sequence List
- Sequence Details (nucleotide view and statistics)
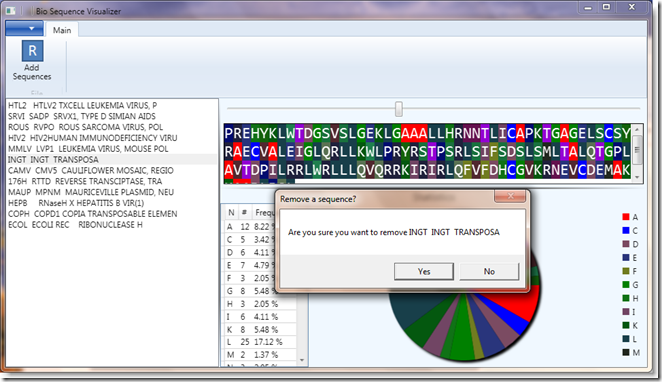
In addition, it uses some of the new Prism v4 features to trigger UI behavior from a ViewModel, this is used when you right-click on a sequence in the sequence list. It drives an action (in the WPF.Behaviors assembly) to display a MessageBox prompt to decide whether to delete the sequence from the list.
Using .NET Bio
The usage of .NET Bio is performed in a few places. First, the Sequence.Data module is where we parse out sequences – this just uses the native parsing capability with one extra twist. If the framework cannot identify the file type itself, we scan the supported extensions to see if we can identify it on our own:
public IEnumerable<ISequence> Load(string filename)
{
var parser = SequenceParsers.FindParserByFileName(filename);
if (parser == null)
{
string extension = Path.GetExtension(filename);
if (!string.IsNullOrEmpty(extension))
{
parser = SequenceParsers.All.FirstOrDefault(sp => sp.SupportedFileTypes.Contains(extension));
}
}
return parser != null ? parser.Parse() : null;
}
In addition, we use the SequenceStatistics class to analyze the sequence that has been selected. This is done in the SequenceStatisticsViewModel class:
public IEnumerable<Tuple<char, double>> Statistics
{
get
{
if (_sequence != null)
{
SequenceStatistics stats = new SequenceStatistics(_sequence);
foreach (byte symbol in _sequence.Alphabet)
{
yield return Tuple.Create((char) symbol, stats.GetFraction(symbol));
}
}
}
}
Reading the code
I tried to keep to fairly simple coding so it could be easily understood – in addition, all the classes and methods are heavily commented for clarity. Here’s the entire project:
This includes the source code, pre-built binaries and a sample .fasta file you can load. You should be able to load other data files as well.
The solution has the following projects:
| Project | Description |
|---|---|
| Contracts | This project contains the interfaces and shared types which all of the other projects need access to. |
| SequenceData | This project contains the sequence loading service – it exposes this service via Prism to interested parties. There is no UI in this project. |
| SequenceAnalyzer | This project is the primary UI host – it initializes the Prism bootstrapper and creates the UI regions where everything is placed. |
| SequenceDetailsUI | This project is generates the sequence detail UI elements – the nucleotide display and the statistics charts. It relies on a 3rd party, open-source charting toolkit. |
| SequenceLoaderUI | This project contains the loader UI (OpenFileDialog) and the currently loaded sequences list which drives selection for the application. |
| SequenceRibbonUI | This project contains the Ribbon docked to the top of the main window. |
| WPF.Behaviors | This project contains an implementation of a MessageBoxPromptAction for Prism to use when we close the application. |
In future blog posts, we’ll look in detail at some of the above elements, and also extend this to include other functionality for analyzing the sequences.